Introduction
Clonal hematopoiesis (CH) refers to non-cancerous clonal expansion of blood cells due to genomic alterations in hematopoietic stem cells. The CH phenomenon observed in people without any hematologic malignancies is particularly called CH of indeterminate potential (CHIP). In a liquid biopsy, variant allele frequency (VAF) of cancer-derived mutations are sometimes indistinguishable from those of CHIP variants. Therefore, CHIP variants could disturb its precise diagnostics. Several studies have suggested ethnic deference of CHIP. However, large Japanese population-based CHIP studies have not been reported yet. We aimed to detect CHIP variants by bioinformatical and mathematical approaches from whole genome sequence (WGS) data provided by Tohoku Medical Megabank (TMM) project, the largest population-based genome cohort study in Japan.
Methods
We used WGS data of 50,316 Japanese individuals who participate in the TMM genome cohort study and analyzed 224,327,538 variants. These genomic data were linked to lifestyle and medical data, including age, sex, past medical history, diet, body weight, smoking habit, depressive mood, and blood and urine testing results.
In general, VAF distribution of CHIP variants is peaked in up to 10-20 percent which should be distinct from those of others (mainly composed of germ-line variants and rare passenger variants). Thus, if representative VAF distributions of CHIP and others was determined, likelihood of CHIP can be estimated for each variant. First, participants with the depth < 20 were excluded for each variant to guarantee the accuracy of VAFs. Second, 223 variants reported as CHIP in TOPMed project, a large-scaled WGS study in the US, were referred as the positive control. Third, the probability density curves of VAF frequency in the positive control and other variants were estimated by the Kernel density estimation (KDE). Last, a ratio of the density value of the positive control to those of other variants were calculated for each specimen and were summarized as a common logarithm value of their geometric mean (called L-index) for each variant. Variants with a high L-index are to be defined as CHIP candidates. The threshold of L-indices was defined as a maximum number where frequency of VAFs ≥ 0.5 does not exceed 1% (finally determined to 0.74).
CHIP variants are classically defined by the following three criteria. (1) Variants detected from peripheral blood cells of people without hematologic malignancies, (2) variants identical to those found in patients with hematologic malignancies, and (3) acquired somatic variants. We found 3,108 variants of 878 genes that were reported in COSMIC, ClinVar, cBioPortal, or MGeND as hematologic malignancy-derived variants (criterion 2). Among them, variants with high L-index were finally selected as CHIP candidates (criterion 3). Common variants detected in more than 1/3 of all participants were excluded because it is not suitable to consider pathogenicity.
Results
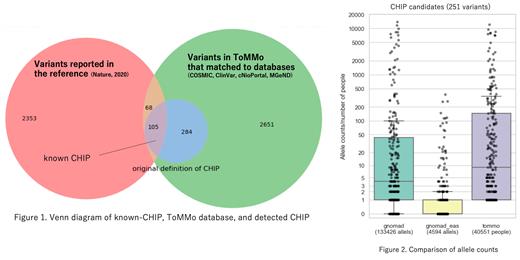
We found 389 variants of 98 genes as CHIP candidates, which included 82 variants (21%) of DNMT3A, 49 variants (13%) of TET2, and 11 variants (3%) of ASXL1. We found newly defined 284 CHIP variants of 94 genes in the Japanese population as well as previously reported 105 variants of 9 genes (Figure 1).
We compared prevalence of these CHIP candidates in the world-wide population. The median allele counts of 251 CHIP candidates registered in the gnomAD were 4 in total population (average 133,426 alleles), 0 in East Asians (average 4,594 alleles), and 9 people in TMM WGS data (average 40,551 people). (Figure 2) Thus, the CHIP candidates were apparently concentrated in the Japanese population.
Conclusions
We systematically detected 389 CHIP candidates from the Japanese WGS database. Our approach matched the classical definition of CHIP, though its biological features are to be evaluated. These candidates were more prevalent in the Japanese population than those in the world-wide population and two-thirds of them were not reported as CHIP in the previous study. Thus, reference-based annotation in liquid biopsies could have overlooked Japanese-specific CHIP variants. We are now evaluating the concordance of the CHIP candidates and true CHIP variants detected in patients with solid tumors. We are also applying our list to other clinical utilities, such as estimating risks of relapse and chemo-resistance of hematologic malignancies.
Disclosures
Tsuchihara:ONO PHARMACEUTICAL Co., Ltd.: Research Funding. Minami:Shionogi: Research Funding; Tejin Pharma: Research Funding; Takeda: Research Funding; Taiho Pharmaceutical: Research Funding; Sumitomo Pharma Oncology: Research Funding; Sanofi: Research Funding; Otsuka: Research Funding; Nippon Shinyaku: Research Funding; Nihonkayaku: Research Funding; Kyowa Hakko Kirin: Research Funding; Dainippon Sumitomo Pharma: Research Funding; Asahi Kasei: Research Funding; Taiho Pharmaceutical: Honoraria; Shinogi: Honoraria; Sanofi: Honoraria; Otsuka: Honoraria; Ono Pharmaceutical: Honoraria; Merck: Honoraria; Meiji Seika Kaisha: Honoraria; Lilly: Honoraria, Research Funding; Kyowa Hakko Kirin: Honoraria; Eisai: Honoraria, Research Funding; Daiichi Sankyo: Honoraria, Research Funding; Chugai Pharma: Honoraria, Research Funding; Bayer: Honoraria, Research Funding; Abbvie: Honoraria; Pfizer Japan Inc.: Honoraria; Bristol-Myers Squibb Company: Honoraria; Takeda: Honoraria; Novartis Pharma KK: Honoraria. Yamashita:Boehringer Ingerheim Japan: Research Funding.